 Back to projects
Back to projects
Assembly and haplotyping of intra-host viral populations from NGS reads
- Download: You can download the latest version of the software here
- References: P. Skums, A. Artyomenko, O. Glebova, S. Ramachandran, I.I. Mandoiu, D. S. Campo, Z. Dimitrova, A. Zelikovsky, and Y. Khudyakov, Computational Framework for Next-Generation Sequencing of Heterogeneous Viral Populations using Combinatorial Pooling , Bioinformatics, 31(5): 682-690
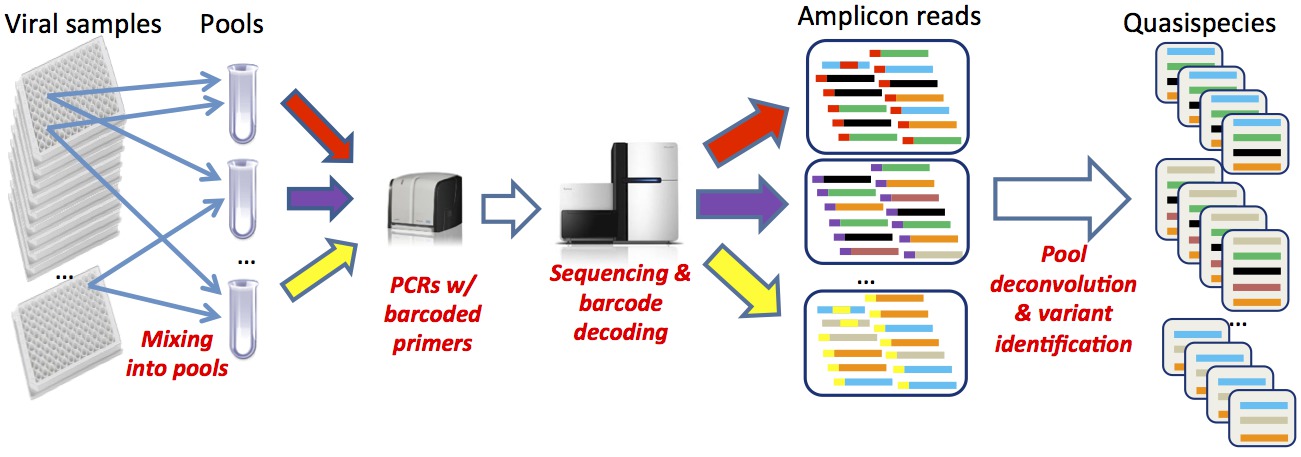
Next-generation sequencing (NGS) allows for analyzing a large number of viral sequences from infected patients, providing an opportunity to implement large-scale molecular surveillance of viral diseases. However, despite improvements in technology, traditional protocols for NGS of large numbers of samples are still highly cost- and labor-intensive. One of the possible cost-effective alternatives is combinatorial pooling. Although a number of pooling strategies for consensus sequencing of DNA samples and detection of SNPs have been proposed, these strategies cannot be applied to sequencing of highly heterogeneous viral populations.
We developed a cost-effective and reliable protocol for sequencing of viral samples, that combines NGS using barcoding and combinatorial pooling and a computational framework including algorithms for optimal virus-specific pools design and deconvolution of individual samples from sequenced pools.
